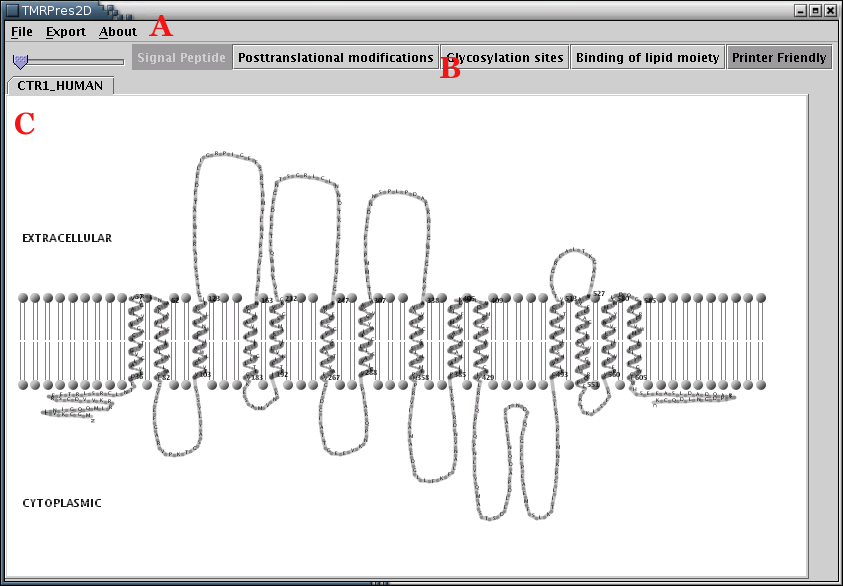
Illustration
1) The main application interface consists of 3 areas. Area 'A' is
the Menu Bar, area 'B' is the Tool Bar and area 'C' is the
Representation Panel. This screenshot corresponds to
printer-friendly output.
TMRPres2D – High quality visual representation of transmembrane protein models
Version 0.91
User's manual
Ioannis C. Spyropoulos, Theodore D. Liakopoulos, Pantelis G. Bagos
and Stavros J. Hamodrakas
Department of Cell
Biology and Biophysics, Faculty of Biology,
University of Athens,
Panepistimiopolis, Athens 157 01, Greece
The Application Workspace
The Application Workspace is divided into 3 parts[Illustration 1]:
The Menu Bar, where the user can handle the basic input/export procedures.
The Tool Bar, where you can accomplish non-disfiguring scaling of the image and adjust various appearance options.
The Representation Panel, where the actual image resides and all user interaction takes place.

Illustration
1) The main application interface consists of 3 areas. Area 'A' is
the Menu Bar, area 'B' is the Tool Bar and area 'C' is the
Representation Panel. This screenshot corresponds to
printer-friendly output.
The Menu Bar
The Menu Bar is always visible and consists of three menus:
File
Export
About
'File' menu
The 'File' menu provides various available methods of data input to the application. The 'Input' submenu includes 4 options, corresponding to different data sources:
SwissProt: SwissProt entries may be loaded from the local disc [Illustration 2], if available, or pasted into the appropriate text area [Illustration 4]. Finally, an entry can be fetched directly from the web, by providing its Accession Number or ID [Illustration 3].
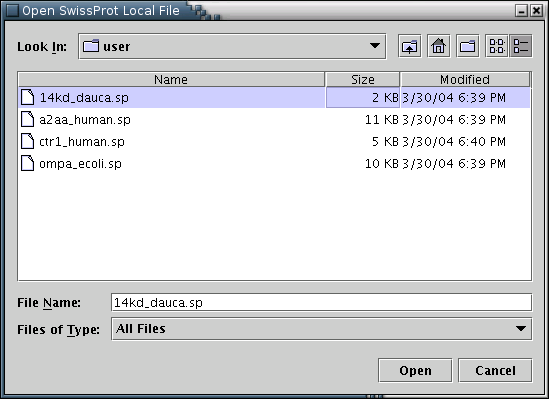 Illustration
2) Window for loading a SwissProt entry from a local disc. The user
has to locate and select a file of plain text containing the desired
entry(e.g 14kd_dauca.sp), and press the 'Open' button.
Illustration
2) Window for loading a SwissProt entry from a local disc. The user
has to locate and select a file of plain text containing the desired
entry(e.g 14kd_dauca.sp), and press the 'Open' button.
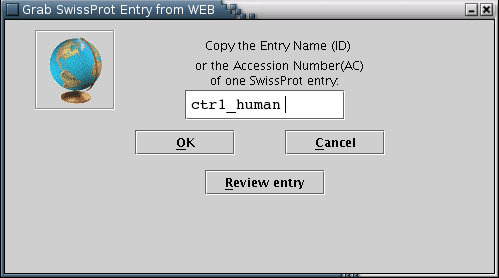
Illustration
3) Input form for downloading a SwissProt entry from the web. After
providing the ID or the Accession Number (eg ctr1_human) of the
entry, the user can press 'OK' for download or 'Review entry' in
order to transfer the entry into the text area form [Illustration
4]
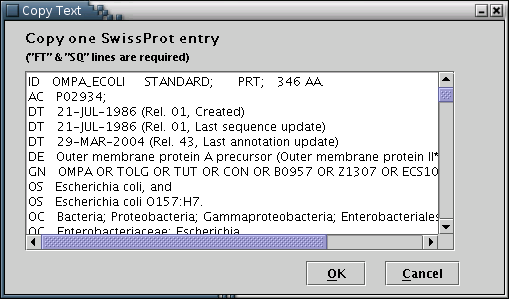
Illustration
4) Input form to paste an SwissProt entry in plain text. After
pasting, the user should press the 'OK' button.
PIR: PIR entries may only be fetched from the web [Illustration 5], by providing the appropriate ID.
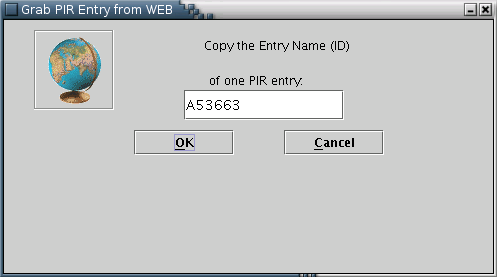
Illustration
5) Input form for downloading a PIR entry from the WEB. After
providing the id (eg A53663) of the entry the user can press 'OK'
for download.
PRED-TMR: Results from the PRED-TMR or PRED-TMR2 prediction algorithms can be used to provide the application with the required information [Illustration 6]. A valid way to provide a PRED-TMR or PRED-TMR2 result, would be to copy and paste the whole webpage containing the output, as provided by the respective web servers (available at http://biophysics.biol.uoa.gr/PRED-TMR and http://biophysics.biol.uoa.gr/PRED-TMR2).
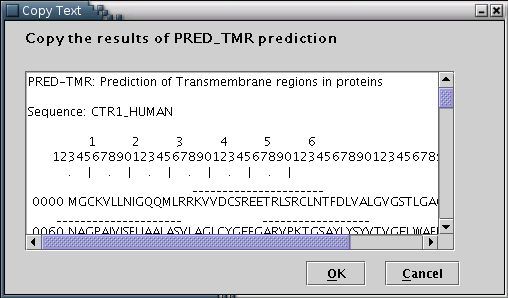 Illustration
6) Input form to paste the PRED-TMR output. Finally, 'OK' button has
to be pressed.
Illustration
6) Input form to paste the PRED-TMR output. Finally, 'OK' button has
to be pressed.
HMMTOP: Results from the HMMTOP prediction algorithm can be used to feed the data input process with the required information[Illustration 7]. A valid way to provide an HMMTOP result, would be to copy and paste the whole webpage containing the output, as provided by the HMMTOP 2 web server at: http://www.enzim.hu/hmmtop/index.html.
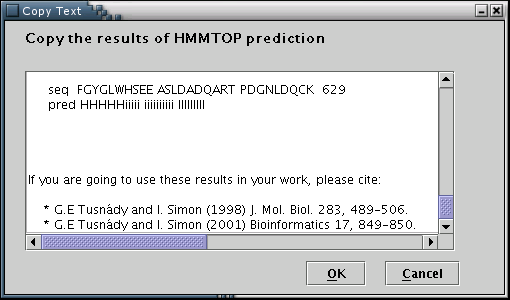
Illustration
7Input form for pasting the HMMTOP output. Finally, 'OK' button has
to be pressed.
TMHMM: Results from the TMHMM prediction algorithm can be used to feed the data input process with the required information[Illustration 8]. A valid way to provide a TMHMM result, would be to copy and paste the whole webpage containing the output, as provided by the TMHMM 2.0 web server at: http://www.cbs.dtu.dk/services/TMHMM/. Please note that the 'Extensive output form' option should have has been selected.
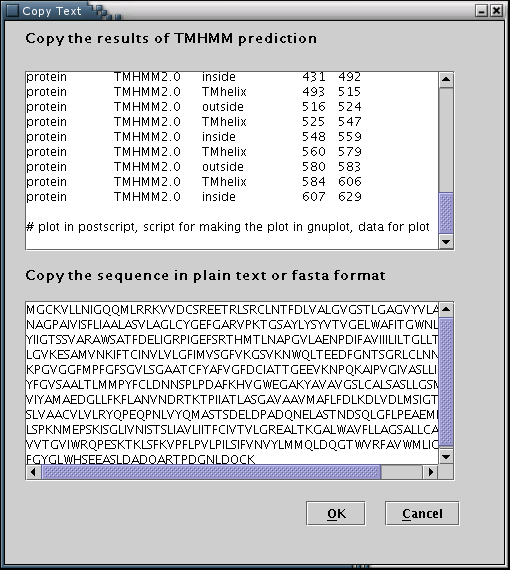
Illustration
8)Input forms for pasting the TMHMM output and the actual sequence.
Finally, 'OK' button has to be pressed.
PRED-TMBB: Results from the PRED-TMBB prediction algorithm (transmembrane topology predictions for beta-barrel outer membrane proteins) can be used to feed the data input process with the required information[Illustration 9]. A valid way to provide a PRED-TMBB result, would be to copy and paste the whole webpage containing a prediction, as provided by the PRED-TMBB server at: http://bioinformatics.biol.uoa.gr/PRED-TMBB .
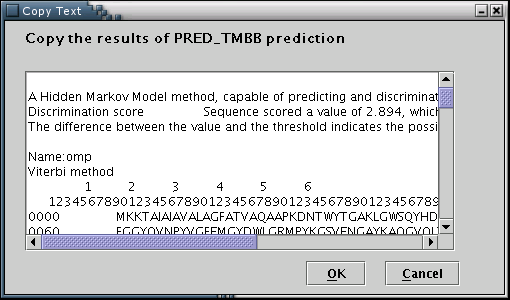
Illustration
9Input form for pasting the PRED-TMBB output. Finally, 'OK' button
has to be pressed.
User defined: Input may be provided by manually filling a special input form [Illustration 10]. The procedure consists of the following steps: You may optionally enter a name for the sequence. The sequence (required) should be entered in the appropriate text area, in plain text or FASTA format. When finished, you should press the 'Apply sequence' button.
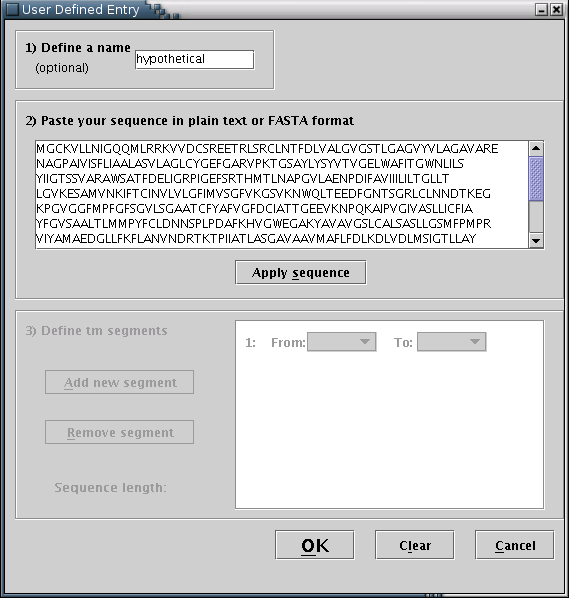
Illustration
10) Input form for filling user defined protein sequence and
transmembrane boundaries.
Consequently, another input area, should be activated [Illustration 11] You may define the transmembrane segments by specifying their boundaries, using the pull down menus provided, named 'From' and 'To'. A new segment should be added each time the 'Add new segment' button is pressed. You can remove the last segment by pressing the 'Remove segment' button once. For each transmembrane segment, a number on the left (colored blue) serves as an index, while a number on the right (colored green) shows its length.
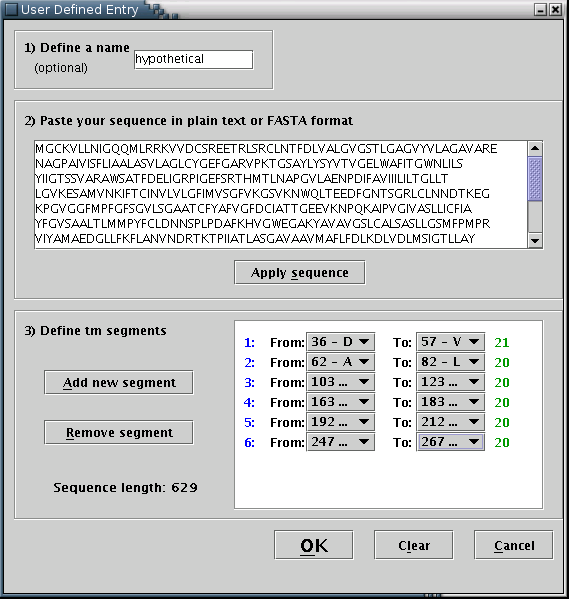
Illustration
11) Input form for filling a user-defined protein sequence along
with its transmembrane boundaries. By pressing the 'Clear' button,
all input fields are reset. By pressing the 'OK' button, the
provided data will be visualized.
'Export' menu
The 'Export' menu provides the option of saving an image/model of a protein using several vector-based formats, namely Encapsulcated PostScript(.eps), PostScript (.ps), Portable Document Format (.pdf) and Scalable Vector Graphics(.svg). Alternatively, pictures may be exported using the popular bitmap image formats JPEG and PNG.[Illustration 12]. This option is enabled only when an image is available.
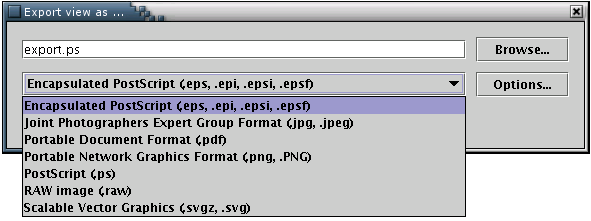
Illustration
12) The export window, from the FreeHEP Java Library. From here one
may select the desirable format and name of the output file or set
some advanced options..
'About' menu
The 'About' menu contains information about the authors and copyright of the application.
The Tool Bar
The Tool Bar should appears after a protein has been loaded [Illustration 13]. It consists of the following elements:

Illustration
13The Tool Bar consists of several toggle buttons and a slider, that
control the appearance of the output.
Scale slider: When you move the knob to the right, the scaling factor is increased, and when you move it to the left, the scaling factor is reduced.
'Signal Peptide' toggle button: If signal peptide data are available, the Signal Peptide toggle button appears enabled. When the button is activated, the signal peptide of the protein appears as a magenta-colored chain.
'Post-translational modifications' toggle button: When you activate this button, special symbols [Illustration 14] should appear under residues for which post-translational modification data are available.
'Glycosylation sites' toggle button: When you activate this button, glycosylation symbols [Illustration 14] should appear above residues for which glycosylation data are available.
'Binding of lipid moiety' toggle button: When this button is activated, lipid binding symbols [Illustration 14] should appear at known lipid-binding residues in the chain.
'Disulfide bonds' toggle button: When you activate this button, a bridge [Illustration 15] would appear between any two residues known to be connected with a disulfide bond.
'Annotation' toggle button: When you activate this button, any defined annotation labels [Illustration 16] should become visible.
'Printer Friendly' toggle button: When you activate the Printer Friendly button, several appearance parameters should be modified:
the background color is set to white
the color of the transmembrane segment boundary indexes is set to black
the color of the peptide chain is set to black.
With these changes, you can produce an image capable to be printed exactly as it appears on the screen. When the button is unpressed, the old appearance options and colors are restored.
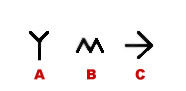
Illustration
14) Symbols for additional sequenee data. A) Glycosylation site,
Lipid binding, Post-translational modification.
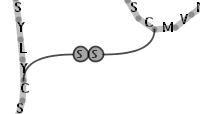
Illustration
15)A symbolic bridge for representing a disulfide bond between two
residues of cystein.
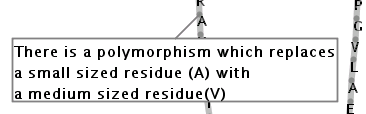
Illustration
16)This text box contains an annotation for a residue. The direction
of the text box, in the above example, is 'bottom left'.
Illustration 17) The 3
potential states of a toggle button. Unselected, Selected and
Disabled, respectively.
Each of the toggle buttons mentioned above,
may appear in one of 3 potential states [Illustration 17]:
Unselected,
Selected
Disabled, whenever the corresponding feature is not present in the input data.

Illustration
17) The 3 potential states of a toggle button. Unselected, Selected
and Disabled, respectively.
The Representation Panel
The Representation Panel should become visible after a protein has been loaded. It contains the actual image. The panel may hold several panes each one containing an image/model. You may switch between models by selecting the corresponding tab [Illustration 18].
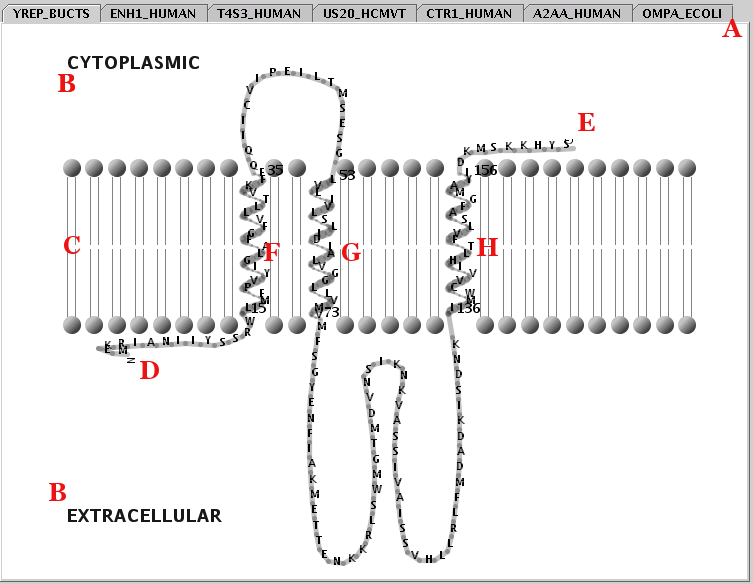
Illustration
18) A view of the Representation Panel. By selecting a tab (eg
YREP_BUCTS) from area A, the appropriate pane appears. The
representation shown ('printer friendly' version), includes the most
common appearance elements: The cellular topology (B), lipid bilayer
(C), transmembrane segment boundaries (black numbers), sequence
termini (D & E), transmembrane alpha-helices (F, G & H),
non-transmembrane chains (loose loops).
Additional sequence features, namely signal peptides, post-translational modifications, glycosylation sites and lipid bindings can be handled from the Tool Bar [Illustration 13] and, whenever visible, are marked with special symbols [Illustration 14].
By pressing the pointer's right button while the representation area is focused, a pop-up menu appears [Illustration 19]. Several appearance options may be configured using this menu:
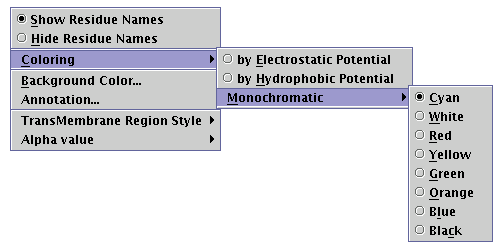
Illustration
19) Semi-expanded pop-up menu.
The 'Show/Hide Residue Names' radio buttons switch toggle the visibility of the aminoacid type names, along the peptide chain. For example, output shown in [Illustration 18] demonstrates the 'Show Residue Names' enabled.
The 'Coloring' submenu carries the coloring options, which may be applied to the peptide chain. These are:
coloring 'by Electrostatic Potential', according to the electrical charge (assuming pH=7), by using blue for positive, red for negative and gray for uncharged residues
coloring 'by Hydrophobic Potential', based on an ad hoc hydrophobicity scale [Illustration 20], by using a gradation from yellow to blue
applying a uniform user-defined color on the protein chain.
 Illustration
20) Coloring by hydrophobic potential.
Illustration
20) Coloring by hydrophobic potential.
The hydrophobicity values assumed for the 20 aminoacid residue types are listed below:
|
Aminoacid |
Hydrophobicity value |
|
Phenylalanine (F) |
0.804 |
|
Isoleucine (I) |
0.734 |
|
Leucine (L) |
0.612 |
|
Tryptophan (W) |
0.582 |
|
Valine (V) |
0.563 |
|
Methionine (M) |
0.407 |
|
Alanine (A) |
0.324 |
|
Cysteine (C) |
0.184 |
|
Glycine (G) |
0.147 |
|
Tyrosine (Y) |
0.073 |
|
Threonine (T) |
-0.129 |
|
Serine (S) |
-0.216 |
|
Proline (P) |
-0.516 |
|
Histidine (H) |
-0.930 |
|
Asparagine (N) |
-0.944 |
|
Glutamine (Q) |
-1.300 |
|
Aspartic acid (D) |
-1.877 |
|
Glutamic acid (E) |
-2.033 |
|
Arginine (R) |
-2.085 |
|
Lysine (K) |
-2.23 |
The 'Background Color' submenu opens a window to choose the background color [Illustration 21].
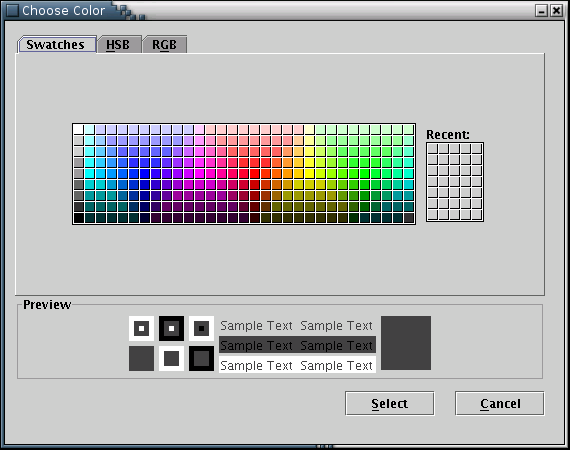 Illustration
21) Window to choose a color for the background.
Illustration
21) Window to choose a color for the background.
The 'Annotation' submenu opens the annotation input form [Illustration 22], from which residue annotations can be added. The desired residue can be selected from the pull-down list. The selection of any residue leads to the appearence of the annotation's message (if available), in the text area. Four radio buttons declare the direction of the box's depiction [Illustration 16]. Any modifications are applied when the 'Submit' button is pressed.
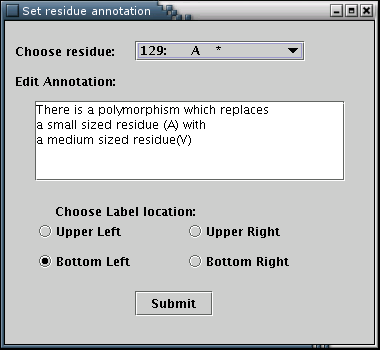
Illustration
22)Window containing the details for adding annotation for a
residue.
The 'TransMembrane Region Style' submenu toggles between two styles of visualizing transmembrane segments, that is, as alpha-helices or as beta-strands. The default style, which is selected everytime a model is loaded, is the helix style.
The 'Alpha value' submenu adjusts the level of transparency of the depicted peptide chain. 'High' alpha values correspond to low transparency, while 'Low' alpha values correspond to high transparency.
By moving the pointer within the representation area while the pointer's left button is pressed, the user can move the image towards a desired direction.