User manual
of cuticleDB
a
relational database of Arthropod cuticular proteins

Christiana
K. Magkrioti, Ioannis C. Spyropoulos,
Vassiliki A. Iconomidou, Judith H. Willis and
Stavros J. Hamodrakas
downloaded from httt://bioinformatics.biol.uoa.gr/cuticleDB
Contents
· Basic Theory
· Data retrieval
Search
in fields
Explaining
the search fields
Name
Taxonomy
Pattern
References
Combined
examples
Gather a set of entries
Protein name
References
Taxonomy
Expression Details
Tissue
Developmental stage
Sequence
Method
Length
Source
Patterns
Signal
Peptide
Function
Putative
Precursor
Fragment
Comments
Exporting to Fasta
Basic Theory
Cuticle is the protective shield of
Arthropods. Its basic constituents are
chitin (the polymer of N-acetylglucosamine) and
proteins. These two macromolecules are
combined to build a complex structure, not only strong, but flexible as
well. The architecture of cuticle is helicoidal and most probably this building plan is
responsible for its extraordinary mechanical, thermal and physiological
properties. In this helicoidal
structure, chitin is in the form of crystalline filaments and proteins play the
role of the matrix.
Although chitin is a simple polysaccharide, the second constituent of cuticle, proteins, present a great variety. Dozens of different proteins may appear in the cuticle of a single organism on a single moment. These proteins may differ between developmental stages or, most commonly, their quantitative distribution may change. It seems that the combination of specific proteins at specific quantities is unique for each region of cuticle, for each developmental stage and for each species.
However,
in this variety some common features can be found. All structural cuticular
proteins are small (with an average length of 100-200 aminoacid
residues), they lack cysteine
and share several characteristic motifs.
These motifs may be small (e.g 3 residues) and
repetitive or large (68 residues) occurring only once (3,4). The most well known motif was recognized by Rebers and Riddiford in 7 cuticular proteins and was called “R&R motif” (1). As more sequences became available, this
motif was widely recognized (2). The
initial consensus was 35 amino acids long, but now encompasses 68 residues as
sequence similarity was recognized at its amino-terminus and the carboxy-teminus was shortened (3,4). This
68 amino acid region, named the "extended R&R consensus" is what
is recognized by PF00379, the Pfam motif for chitin
binding of Arthropod cuticle (3, 4, 5). This "extended R&R consensus"
is most probably dominated by b-sheet structure (6,7).
Three types of the "extended R&R consensus" have been found: RR1, RR2 and RR3 (8, 9). The two main types are RR1 and RR2, which presumably appear in proteins from soft and hard cuticles, respectively (8). Very few cuticular protein sequences show the existence of RR3 (9). Actually, the secondary structure predicted for these two types RR1 and RR2, verifies the fact that they appear in regions of cuticle with different properties (7).
Scope of the database
cuticleDB is the first database of Arthropod cuticular proteins.
The goal of its constructors was the collection of all cuticular protein sequences that have appeared to date and
their detailed and correct annotation.
The better the organisation of the data, the easier the work will be for
researchers dealing with cuticle and structural proteins in general.
It is hoped that, this database will be
of help to genome annotators in the near future when more arthropod genomes
will become available. Furthermore, it
is hoped that, detection of common properties of these proteins, as well as
recognition of important differences that are responsible for cuticle’s
complexity and important functions will be facilitated by its existence. The database will be updated at regular
intervals.
Data Retrieval
From the 'Data Retrieval'
page, you can retrieve one or more entries.
This retrieval can be done in two ways:
A) Search
in fields
B) Gather
a set of entries
A) Search in fields
By 'Searching in fields' the user can
search for any text in the fields of his/her preference. The user can enter any word/expression in one
or more of the available boxes under the name: 'Name', 'Taxonomy', 'Pattern' and 'References'.
Each expression may contain:
i)
Text
terms to be searched for,
ii)
Parenthesis
'(' ')' which groups one or more sub-expressions,
iii)
Operator
'&' for AND, which combines two (or more) sub-expressions in
a single field and gives the user the opportunity to search for entries that
satisfy all sub-expressions.
iv)
Operator
'|' for OR, which combines two (or more) sub-expressions in a
single field and gives to the user the opportunity to search for entries that
satisfy at least one of the sub-expressions.
v)
Operator
'!' for NOT, which can only be used at the beginning of an
expression and does not connect two sub-expressions. It provides to the user the opportunity to
search for entries that necessarily do not satisfy the expression after the
'!'.
vi)
Operator '&!' for AND NOT, which
combines two (or more) sub-expressions in a single search field and gives to
the user the opportunity to search for entries that satisfy only the sub-expression in the left of the operator.
Expressions in separate search fields are
combined with the AND operator, so every entry of the result set will satisfy
the expressions of all the search fields the user has chosen.
Note: The special characters '(' and ')' can be searched literally using first '\', eg: \(
Explaining the search fields
The four available search fields are
shown as four boxes named: 'Name', 'Taxonomy', 'Pattern' and 'References'.
Each box (apart from the 'Pattern' box) corresponds to a certain field of
the database entries.
-
'Name'
corresponds to the field
'Protein name' of an entry. The text
written by the user (query) is searched against the information included in all
cuticleDB entries under this field. It is not necessary to have an exact match
between the query and the complete name of a protein. The only condition for a positive result is
that the query is part of the protein name of at least one entry. Moreover, the search is not case sensitive,
so the user does not have to worry about uppercase and lowercase letters.
Examples:
1. If the user wants to retrieve a protein that he/she has found in the literature under the name LCP65Aa, the query should be this name exactly.
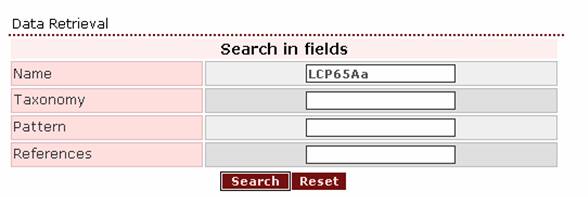
The result page will be like this:
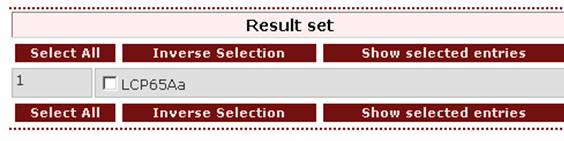
In case the user wants to retrieve all
proteins from this multigene family, the appropriate
query would be ‘LCP65’. That is:
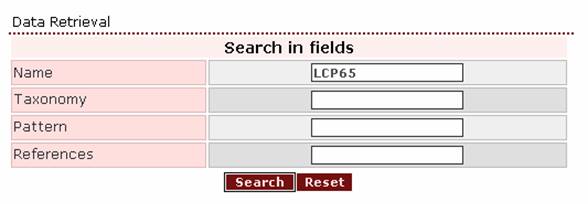
and the result page will be:
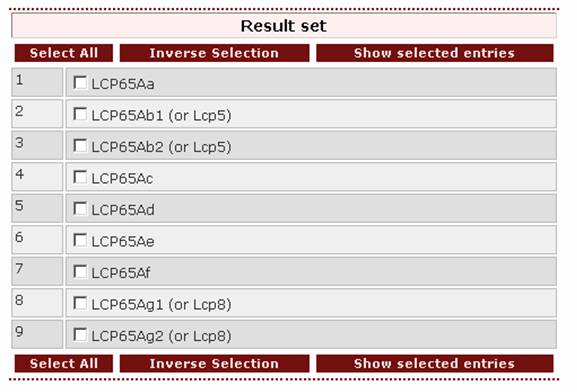
As a general advice for someone who wants to retrieve the entry of one specific protein, he/she should set as a first query the protein name found in the literature. In case the search is negative, the user should try to shorten the query. For example, searching for the Manduca sexta protein with the query ‘MsCP27’ or ‘mscp27’ (as the search engine is case insensitive) does not give a result.
Negative result page:
However, searching with the query
‘cp27’ it produces a positive result.
Positive result page:
2. If the user wants to retrieve one or more proteins that are expressed in the larval cuticle, a proper query would be ‘LCP’.
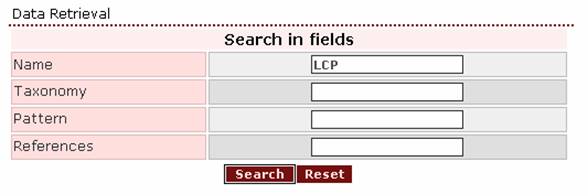
The top of the corresponding result page is:
- 'Taxonomy' corresponds to the field Taxonomy of an
entry. The query may include the name of
any taxon from the rank of superkingdom
to genus; but, be careful!, not species. The query should match with at least part of
the taxonomy field in one or more entries.
Examples:
1.
If
the user wants to retrieve all cuticular proteins
from the genus Drosophila, he/she should use this name as a query in the
Taxonomy box.
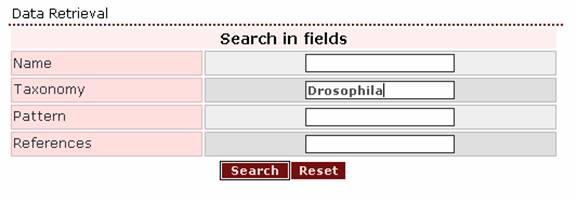
The top of
the result page is:

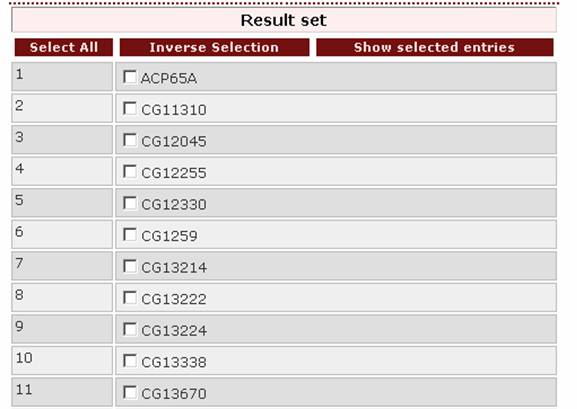
Setting half of the genus’ name as a query: ‘dros’ will also work.
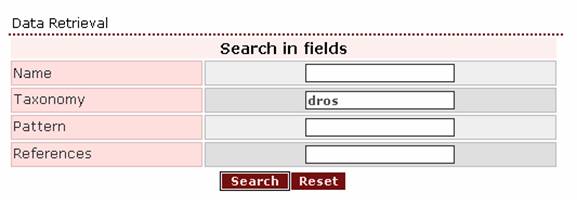
The result page will be the same:
2.
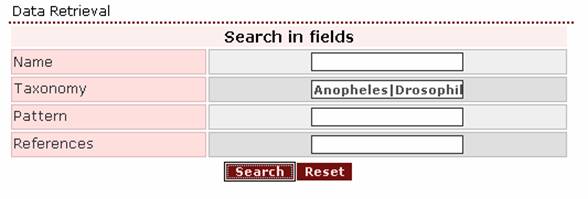
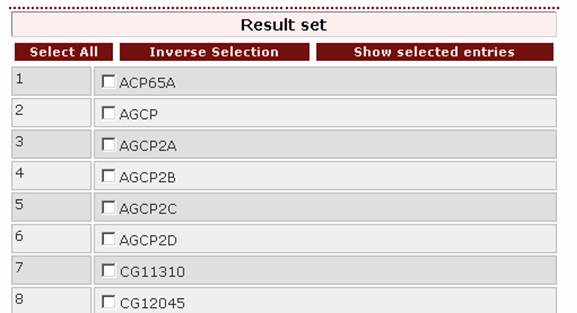
If
the user wants to retrieve all protein entries assigned to Anopheles and Drosophila, the proper query is
‘Anopheles | Drosophila’.
The top of the result
page is:
3.
If
the user wants to retrieve all proteins from the order Lepidoptera apart from
those of the genus Hyalophora, then an appropriate
query is ‘Lepidoptera &! Hyalophora’.
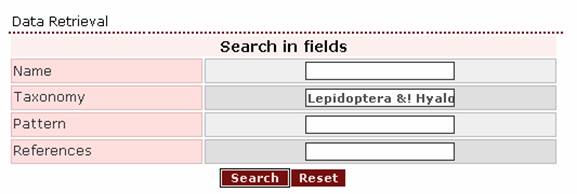
The top of the result page is:
4.
If
the user wants to retrieve all protein entries of cuticleDB,
an easy way is to set as query the word ‘Arthropoda’
in the Taxonomy search field.

The bottom of the result page shows the exact total number of cuticleDB entries (445):
-
'Pattern'
does not correspond to
an entry field. In this box the user can
enter whichever motif he/she pleases, as long as this includes valid
characters. Valid characters are:
|
The 20 symbols of the one-letter
amino-acid code, |
|
The letter X for unknown aminoacid positions, |
|
The characters ' [' and '] ' in order
to enclose two or more choices for the same aminoacid
position, and |
|
The character '|' in order to
separate these choices. |
Examples:
- If the
user wants to search all cuticleDB proteins for
the pentapeptide ala ala ? phe
his, the right query-pattern would be ‘AAXFH’. Of course, ‘aaxfh’
would also be valid.
The
result page is:
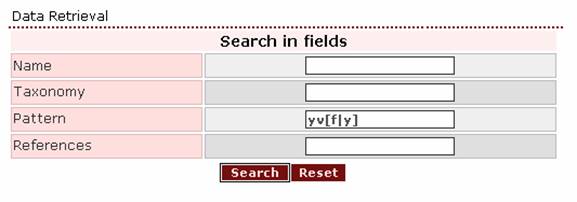
- If the user wants to search for the pattern tyr val
(phe / tyr),
the right query-pattern would be ‘yv[f|y]’.
The
top of the result page is:
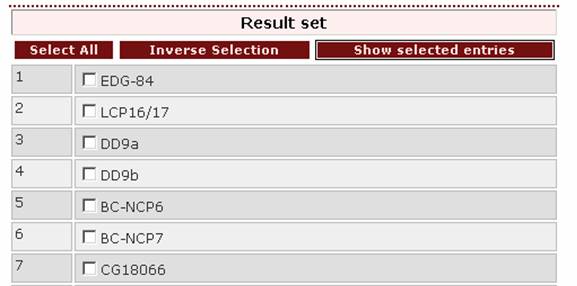
Note: The query ‘yv(f|y)’ is not valid.
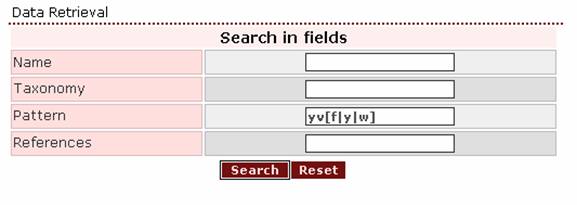
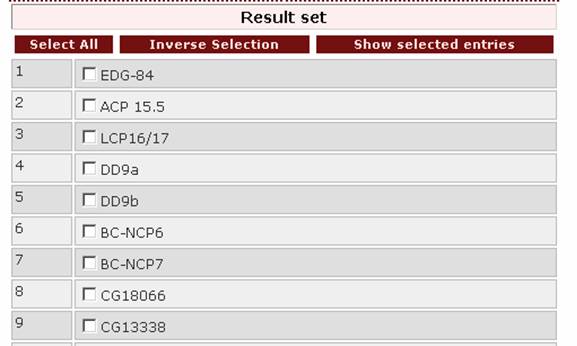
- If the user wants to search for the pattern tyr val
(phe/tyr/trp), the appropriate query-pattern is ‘yv[f|y|w]’.
The top of the result page is:
Attention!: The patterns
shown in the entries of the result set are the preselected
patterns that the constructors of the cuticleDB have
located. The query-pattern of the user,
along with its start and end positions, will be shown only if the user searches
for this query-pattern again, independently in each entry. The query in each entry is made through the
box:
![]()
This is found in the field Patterns:
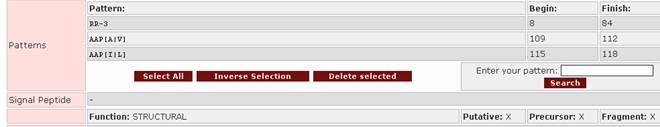
-
'References'
corresponds to the entry
field References. The query may
be one of these codes: 'Entrez accession number', 'Entrez gi', 'Uniprot
AC', 'FlyBase ID', 'Ensembl
ID' or 'Pfam'. (InterPro codes in the near future). The query is searched against the various
database codes included in the field References of all entries.
Examples:
- If the user wants to retrieve
the entry of a protein coded P81581 by Uniprot,
the proper query is this exact code in the References search field.
The result page is:
- If the
user wants to retrieve entries of proteins he/she has found in several
other databases, for example both Uniprot and Entrez, the appropriate query would combine all the
different codes e.g. ‘P81584 | P81585 | 102879 | 113012’, where the first
two are Uniprot Accession Numbers and the last two
are Entrez GenInfo
Identifiers.
The result set is:
Combined examples
- If the user wants to retrieve
all Anopheles proteins that bear the motif GGLG, then ‘Anopheles’ should be written in
the Taxonomy search field and ‘GGLG’ in the Pattern search
field.
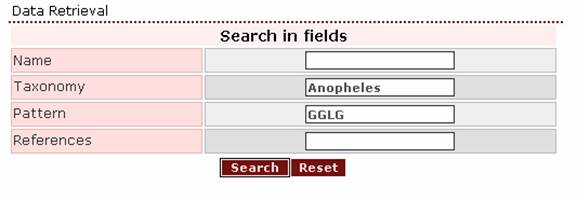
The result
page is:
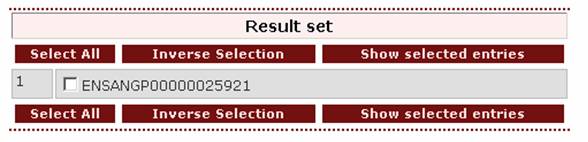
- If the
user wants to retrieve all Anopheles proteins that were not taken from the
Anopheles gambiae genome project, ‘! ensangp’ should be written in
the Name search field and ‘Anopheles’ in the Taxonomy search
field.
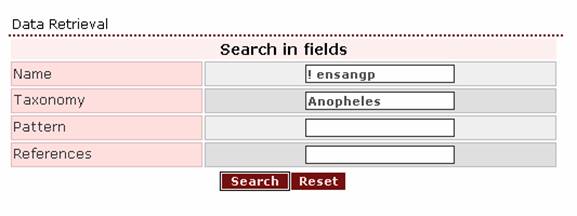
The result page is:
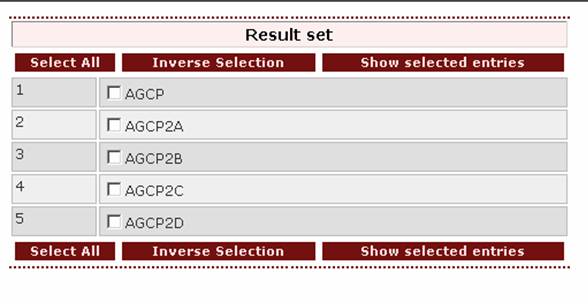
B)Gather a set of entries
Gathering a set of entries is another way for retrieving data. The user can retrieve all database entries referring to proteins that derive from the same species or proteins that share a common pattern in their sequence. Contrary to the patterns in the “Search in fields”, patterns in the “Gather a set of entries” are pre-determined by the creators of the database. These patterns have been reported in the field Patterns of the detailed view of an entry.
Examples:
1. If the user wants to retrieve all entries describing proteins of the species Homarus americanus, this species should be selected from the pull-down list in the “Search by Species” box.
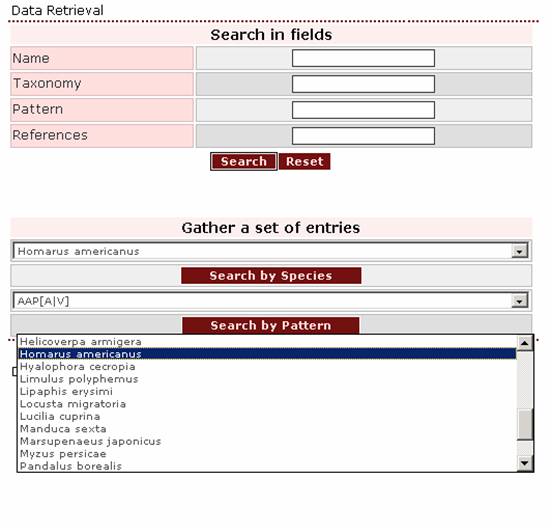
Clicking on the ‘Search by Species’ button the result page appears:
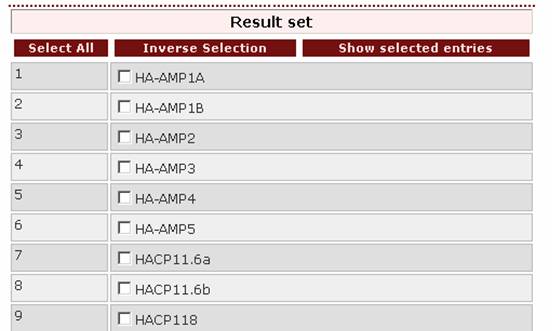
2. If the user wants to retrieve all proteins bearing the RR1 pattern, this pattern should be selected from the pull-down list in the “Search by Pattern” box.
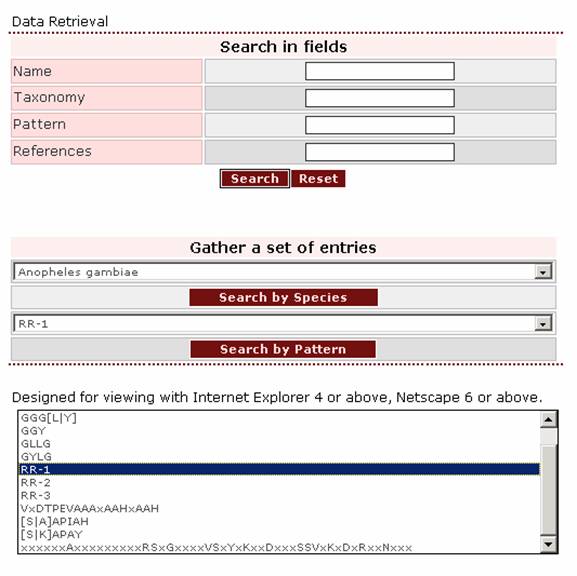
Result set
After querying the database, a result set will appear. From this set the user can select the entries to be viewed by ticking the boxes on the left of the proteins:
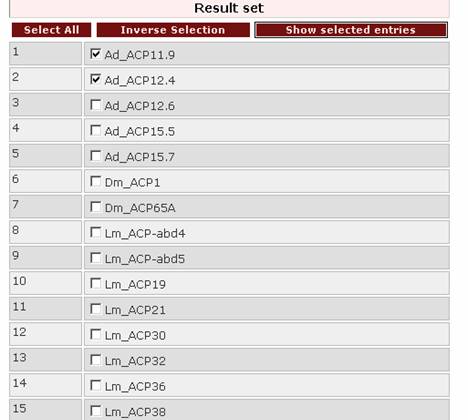
and then clicking on
![]() .
.
Additionally, by clicking on the button 'Select all' all the entries of the result set are selected. By clicking on the button 'Inverse selection' all unchecked boxes are selected.
Either way, all entries
chosen will appear in a common ‘Protein View' page. The user can navigate in this page using the
expressions An entry of the database is as follows: The available fields in a protein entry
of cuticleDB are: Protein name: Contains the name of the protein. In the majority of the proteins, this name
corresponds to the name given by the scientist who determined its
sequence. This name may derive from the
protein’s molecular mass or from the order in which this protein was obtained. Some names include the first letter of the
genus and the first letter of the species from which the protein was
extracted. The name quite often includes
the first letter of the developmental stage at which the extraction took place,
for example a name may include the letters ACP for Adult Cuticular Protein. However, this protein may not necessarily be
stage-specific; it may just have been found at that stage (3,4). For proteins obtained from
the Drosophila melanogaster or Anopheles gambiae genome project, the identifier attributed by FlyBase or Ensembl, accordingly,
has been held. Therefore, the user may
search for names of the type: ·
LCP1
(Larval Cuticular Protein 1;where
1 is the order of the corresponding gene in its cluster) ·
LCP-14
(Larval Cuticular Protein 14;
wher 14 is the molecular mass of the protein) ·
TM-F1A
(Tenebrio Molitor
and F1A, which is the order in which the protein was eluted from cation-exchange chromatography) ·
CG15920
(Computed Gene and number given by FlyBase) ·
EDG-91
(Ecdysone Dependant Gene and
number given by the researcher). References: Contains codes of the protein in other
databases. These codes act as links, as well.
References are provided for the following databases: Entrez
Protein database (accession number and GenInfo
identifier are available), Uniprot, Interpro, Pfam, FlyBase, Ensembl. Taxonomy:
Contains an
hierarchical description of the organism that expresses the protein. The description includes all the taxa from superkingdom until
genus. The species’ name is provided in
a separate text-box. It should be mentioned that all taxonomic information has been extracted from Entrez Protein Database. Expression Details: Contains information about the
time and spatial distribution of the protein or the corresponding mRNA. The subfield Tissue specificity
concerns the appearance of a
protein in specific areas of cuticle or
the occurrence of the corresponding mRNA in specific tissues. The subfield Developmental stage concerns
the appearance of a protein or the corresponding mRNA in a certain
developmental stage. The subfield Reference
shows the bibliographical source used for this field. AAP[A|V] AAP[I|L] [S|K]APAY [S|A]APIAH GYLG GLLG GGG[L|Y] GGY VxDTPEVAAAxAAHxAAH xxxxxxAxxxxxxxxxRSxGxxxxVSxYxKxxDxxxSSVxKxDxRxxNxxx RR-1 RR-2 RR-3 All information given about
the existence of the RR3 motif has been drawn from literature. A Prosite
pattern that can be considered representative for each of these three motifs
RR1, RR2 and RR3 is given below: RR-1 GSYSYTxPDGxxYxVxYVAD-ENGFQPxGxHLP RR-2 EYDxxPxYxFxYxVxDxxTGDxKSQxExRxGDVVxGxYSLxExDGxxRTVxYTADxxNGFNAVVxxEx RR-3 V-xVxTxYHAQDxLGQxSFGHxxxxQxRxExxDAAGNKxGSYxYVDPxGKVxxxxYVAD-AxGFRVAxx-NLPVxP Please, note
that the RR1 and RR2 Prosite patterns may not be
exactly the same in the protein entries that have been marked as bearing the
RR1 and RR2 patterns, since the existence of the RR1 and RR2 patterns has been
detected in the protein sequences of cuticleDB using
profile HMMs, as explained above. Furthermore, the RR1 pattern (built utilizing
profile HMMs) is actually larger than the one shown
here, but given its diversity, no Prosite
type pattern can be built for the entire length of RR1. Signal Peptide: Shows the start and end position of the signal
peptide. The signal peptides have been
detected using SignalP 2.0 (11). More specifically, the Y score of the Neural
Network prediction was used as a selection criterion and the default threshold,
0.32, was used as cut-off for the prediction.
The protein sequences that were derived from proteins extracted from
cuticle - thus, they had already been secreted and had their signal peptide
cleaved off- do not possess a signal peptide. Comments:
This field may include
information of great diversity, such as the existence of protein isoforms, polymorphic sites or the specific strains from
which a certain sequence was derived. Moreover, clicking on the button our web-server sends a text content with all the sequences of
the selected set in Fasta format. Different web-browsers may manage this text
differently. For example, Windows Internet Explorer asks the user how to handle
it, that is, view it with an associated program or save the content in a file. The user must choose ‘Save’ and then save
the file as type “All files” and with a suitable name e.g. “fasta.txt”. Then he
may view the file with an appropriate text viewer. Notice that programs like
Notepad are not suggested; use WordPad instead. In addition, other web-browsers
like Netscape or Mozilla, display the text content
directly. 1. Rebers, J.E. and Riddiford, L.M. (1988) Structure and expression of a Manduca sexta
larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol.,
203, 411-23. 2. Willis, J.H. (1999) Cuticular
proteins in insects and crustaceans. Am. Zool.,
39, 600-609. 3. Andersen, S.O., Hojrup,
P. and Roepstorff, P. (1995) Insect cuticular proteins. Insect Biochem. Mol. Biol., 25, 153-76. Review. 4. Willis, J.H., Iconomidou, V.A., Smith, R.F.
and Hamodrakas, S.J. (2004)
Cuticular proteins. In:
“Comprehensive Insect Science” L. I. Gilbert, K. Iatrou,
and S. Gill, Eds. Elsevier, 5. Rebers, J.E. and Willis, J.H.
(2001) A conserved domain in arthropod cuticular
proteins binds chitin. Insect Biochem. Mol. Biol.,
31, 1083-93. 6. Hamodrakas, S.J.,
Willis, J.H. and Iconomidou, V.A. (2002) A structural model of the
chitin-binding domain of cuticle proteins.
Insect Biochem. Mol. Biol., 32,
1577-1583. 7. Iconomidou, V.A., Willis, J.H. and Hamodrakas, S.J. (1999) Is beta-pleated sheet the molecular conformation which
dictates formation of helicoidal cuticle? Insect Biochem Mol Biol., 29, 285-92. 8. Andersen, S.O. (1998)
Amino acid sequence studies of endocuticular proteins
from the desert locust, Schistocerca gregaria. Insect Biochem.
Mol. Biol., 28, 421-434. 9. Andersen, S.O. (2000) Studies on proteins in post-ecdysial nymphal cuticle of locust, Locusta
migratoria, and cockroach, Blaberus
craniifer. Insect Biochem.
Mol. Biol., 30, 569-577. 10. Eddy, S.R. (1998) Profile hidden markov models. Bioinformatics,
14, 755-763. 11. Nielsen, H., Engelbrecht, J., Brunak, S. and von Heijne,
G. (1997) Identification of prokaryotic and eukaryotic signal peptides and
prediction of their cleavage sites. Protein
Engineering, 10, 1-6. ![]()
![]() at the upper right corner of each entry.
at the upper right corner of each entry.Detailed view
of an entry
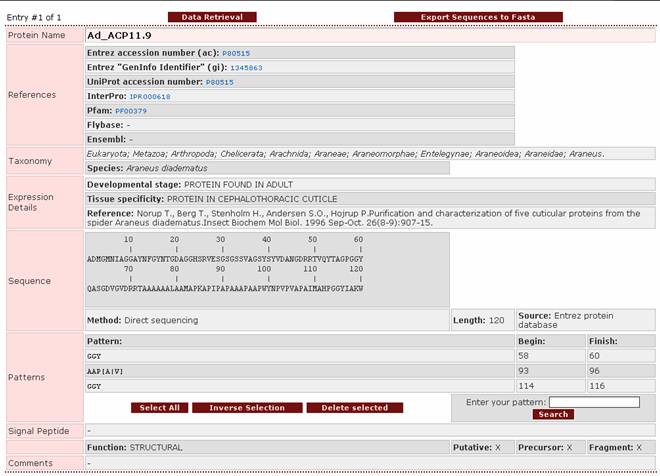
This entry belongs to protein Ad_ACP11.9, which is a
cuticular protein of Araneus
diadematus.
Sequence: Shows the protein sequence, which can also be shown in FASTA format. Aminoacid numbering is shown above the sequence. The subfield Method provides information about the
biochemical method of sequencing. This
method may be direct sequencing of the protein, conceptual translation of the
protein’s mRNA or cDNA or both (for example a protein
sequence may have been derived from conceptual translation of a cDNA and then checked by partial direct sequencing of the
protein). The subfield Length
shows the number of aminoacid residues in the protein. The subfield Source shows the protein
database from which the
sequence was obtained.
Patterns: Shows all the patterns that have been found in the protein sequence,
together with their start and end position.
The patterns that have been searched for were selected from the
literature. They are shown on the
following table:
All the above patterns, apart from RR1, RR2 and RR3,
were located for each entry, utilizing a home-made tool, specifically created
for seeking motifs of the Prosite
type. The RR1 and RR2
patterns were located, utilizing Profile Hidden Markov Models that were created
specifically for this database (unpublished), using the HMMER
software package (version 2.3.2) (10).
Function: Shows the function of the protein. All proteins in the current version of cuticleDB are structural.
This field will be of importance when proteins of other functions will
be included as well.
Putative: Proteins are classified as putative cuticular proteins unless
at least one of the
following criteria has been met. Criteria are: complete or partial
amino acid sequence
data from proteins isolated from cuticle, or an antibody raised against
a specific protein
reacts with cuticle or with proteins isolated from cuticle, or an
antibody raised against a
cuticle extract reacts with a translation product from a specific cDNA,
or a specific
mRNA has been detected in cuticle secreting epidermis.
Preliminary:
Indicates that the sequence based on genome annotation or single pass sequencing appears to be incomplete or incorrect or has been designated by the annotators as preliminary.
Fragment: Gives information about whether the whole sequence or just a fragment of a certain protein
is provided.
Exporting to Fasta
![]()
References