|
 |
Research Activities: Bioinformatics |
|
| top |
|
| The Bioinformatics group of our
lab is mainly focused on the development
of algorithms that predict the structure, function and interaction of
proteins. These algorithms frequently have genome-wide applications.
More specifically: |
|
| Prediction of protein structure,
function and interactions |
| Our
research is mainly focused on developing algorithms that are capable
of predicting the structure, function and interactions of proteins. We
are, in particular, focusing our efforts on the topology
prediction of transmembrane proteins, either alpha-helical ones
(PRED-TMR,CoPreThi, orienTM, waveTM) or
beta-barrels (PRED-TMBB, ConBBPred). Furthermore, we
developed algorithms for structural and/or functional classification
of proteins readily available for genome-wide applications. Such
applications are available for structural classification of proteins
(PRED-TMR2, PRED-CLASS, MCMBB) and for
classification of the G-protein Coupled Receptors into families
(PRED-GPCR), or according to the G-protein coupling
specificity (PRED-COUPLE, PRED-COUPLE2). Towards these goals, we use sophisticated mathematical and computational methods ranging from various methods of statistical analysis, to Neural Networks, Support Vector Machines, Dynamic Programming and Hidden Markov Models. |
|
| Development and annotation
of biological databases |
| The
knowledge expertise of our team, in combination with the
sophisticated tools that we develop, enables us to build curated,
expert, knowledge-based, relational biological databases. These
databases may be used either by bioinformaticians or by molecular
biologists. We have been focused, until now, mainly on biologically
interesting classes of proteins that, either are not fully annotated
in publicly available databases, such as the Outer Membrane Proteins
(OMPs) of Gram-negative bacteria and the insect cuticle proteins
(cuticleDB), or to classes, with important functional roles that involve protein-protein interactions (gpDB, mpMoRFsDB). |
|
| Sequence alignment, Periodicities, and low complexity regions discovery |
| Our
research team focuses also on discovering hidden periodicities in
protein and DNA sequences, using either the Fourier Transform (FT)
or the Wavelet Transform (waveTM). We are also studying the
low complexity regions in protein and nucleic acid sequences, and we
developed algorithms for filtering such sequences (CAST). Using
pairwise alignments, we implement algorithms for clustering
large datasets and create non-redundant sets (NON-RED). Recently,
research has been focused on improving the pairwise and
multiple alignment algorithms quality, by either incorporating
structural or evolutionary information (unpublished). Furthermore,
tools for the represantation of membrane protein structure were also
developed (TMRPres2D), as well as tools capable of visualizing
gene-product functional and structural features in genomic datasets
(GeneViTo). |
 |
|
|
|
 |
Research Activities: Biophysics |
|
| top |
| The
scientific research interests of our laboratory are also oriented
towards the field of structural biology and molecular biophysics.
More specifically, we are focused on: |
|
| Structural studies of peptide-analogues parts of silkmoth chorion proteins, as novel self-assembled polymers with amyloid properties. |
| Chorion
is the major component of silkmoth eggshell. More than 95% of its
dry mass consists of proteins that have remarkable mechanical and
chemical properties protecting the oocyte and the developing embryo
from a wide range of environmental hazards. Synthesized
peptide-analogues of parts of chorion proteins fold and self-assemble
forming amyloid-like fibrils, under a wide variety of diverse
conditions. This raises the question whether chorion is a natural,
protective amyloid. The folding and self-assembly mechanisms of
these peptides are being extensively studied utilizing electron
microscopy (negative staining and shadowing), X-ray diffraction,
FT-Raman, ATR FT-IR and CD spectroscopy and computer modelling.
Principles that govern the self-assembly of proteins into
amyloid-like structures are being unravelled and may be important in
a variety of pathological cases in amyloidoses like Alzheimer’s, transmissible spongiform encephalopathies (mad-cow disease, Creutzfeld-Jacobs, prion diseases etc.), type II diabetes etc. |
|
| Studies of protein-chitin
interactions for the formation of arthropod cuticle. |
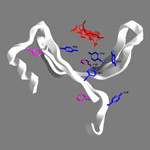 |
Cuticle
is a complex, bipartite structure, composed mainly of proteins and
chitin, which provides protective, locomotive and structural
functions important for arthropod survival. Chitin-protein
interactions are studied utilizing X-ray diffraction, FT-Raman, ATR
FT-IR and CD spectroscopy of natural and specially treated samples.
Cuticle protein sequence alignment, secondary structure prediction,
computer modelling and docking studies are also used. Recently, a
relational cuticle protein database (cuticleDB), was also
constructed in our lab and is freely provided through the internet.
Also, recently, a model for the structural proteins both of hard and
soft cuticles has been proposed and guides our efforts towards
unravelling the molecular principles that dictate cuticle overall
molecular architecture. |
|
| Structural and self-assembly studies of
fibrous proteins, which form structures of physiological importance
like silkmoth and fish chorion. |
 |
Silkmoth and fish chorion is a helicoidal composite (biological analogue of a cholesteric liquid crystal) of protein fibres embedded in a protein matrix. The principles governing the self-assembly of chorion protein molecules into helicoidal proteinaceous extracellular structures are being studied. X-ray diffraction, FT-Raman, ATR FT-IR spectroscopy, electron microscopy and computer modelling are the main techniques used to achieve this goal. |
|
|
| Structural studies of protein molecules that play significant roles in cell functions, through determination of protein structures (and as complexes with small organic compounds) utilizing X-ray crystallographic methods. |
| Research is carried out on the enzyme Dihydrofolate Reductase (DHFR), which participates in a biological pathway that leads to the formation of thymine which is essential in DNA biosynthesis and various aminoacids. Blockade of the action of this enzyme leads to cell death. In the context of anticancer and antimicrobial research, DHFR has extensively been studied. Our goal is to study the interaction of DHFR with small organic compounds, inhibitors of the enzyme in order to design compounds appropriate for clinical treatment. |
 |
Furthermore,
efforts are centered on the study of Concanavalin A (Con A), which is
a representative member of the lectin class of plant proteins. It
generally exhibits specificity for saccharides containing a-D-mannose
or a-D-glucose residues, but it may also bind oligosaccharide
sequences lacking these units. Con A has specific biological
activities which depend on its binding to cell surface receptors. It
agglutinates cells transformed by oncogenic viruses, inhibits growth
of malignant cells in animals, and exhibits mitogenic activity.
Although the exact biological role of Con A still remains unknown,
its specific saccharide-binding properties make it an ideal object
for the study of protein–saccharide interactions. |
|