

The 'TransMembrane
protein Re-Presentation
in 2 Dimensions'
tool, automates the creation of uniform, two-dimensional, high analysis
graphical images/models of alpha-helical or beta-barrel transmembrane
proteins. Protein sequence data and structural information may be
acquired from public protein knowledge bases, emanate from prediction
algorithms, or even be defined by the user. Several important
biological and physical sequence attributes can be embedded in the
graphical representation.
Our method offers several characteristic unique features:
Our method offers several characteristic unique features:
 Visualization
of both alpha-helix and beta-barrel transmembrane proteins.
Visualization
of both alpha-helix and beta-barrel transmembrane proteins.
 'One-step' data input procedure, including easy querying of the public databases
and incorporation of output from various popular prediction tools available from the Web.
'One-step' data input procedure, including easy querying of the public databases
and incorporation of output from various popular prediction tools available from the Web.
 Accurate dimensions of the molecule elements, with adjustable scaling.
Accurate dimensions of the molecule elements, with adjustable scaling.
 Representation
of any available biological and physical attributes, using chromatic
and symbolic variations (e.g. disulfide bridges, signal peptide).
Representation
of any available biological and physical attributes, using chromatic
and symbolic variations (e.g. disulfide bridges, signal peptide).
 Ability to easily incorporate custom, user-defined annotations for any aminoacid residue.
Ability to easily incorporate custom, user-defined annotations for any aminoacid residue.
 Wide variety of supported vector and bitmap-based image formats.
Wide variety of supported vector and bitmap-based image formats.
 Integrated
graphical user interface.
Integrated
graphical user interface.
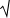 Intelligible
and elegantly styled depiction.
Intelligible
and elegantly styled depiction.Please cite as:
TMRPres2D: high quality visual representation of transmembrane protein models Bioinformatics. 2004; 20: 3258-3260.
 biol.uoa.gr
biol.uoa.gr